 Image credit: G. Reddy
Image credit: G. Reddy
Abstract
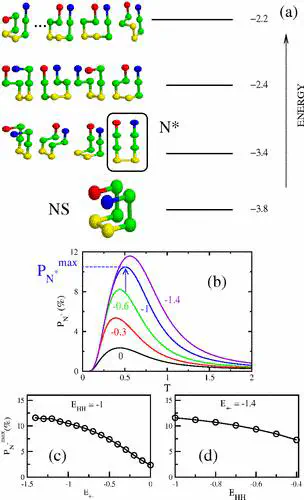
Using lattice models we explore the factors that determine the tendencies of polypeptide chains to aggregate by exhaustively sampling the sequence and conformational space. The morphologies of the fibril-like structures and the time scales ( τ fib ) for their formation depend on a balance between hydrophobic and Coulomb interactions. The extent of population of an ensemble of N ∗ structures, which are fibril-prone structures in the spectrum of conformations of an isolated protein, is the major determinant of τ fib . This observation is used to determine the aggregating sequences by exhaustively exploring the sequence space, thus providing a basis for genome wide search of fragments that are aggregation prone.
Type
Publication
Phys. Rev. Lett., 2010, 105, 218101