Thermodynamics and Kinetics of Single-Chain Monellin Folding with Structural Insights into Specific Collapse in the Denatured State Ensemble
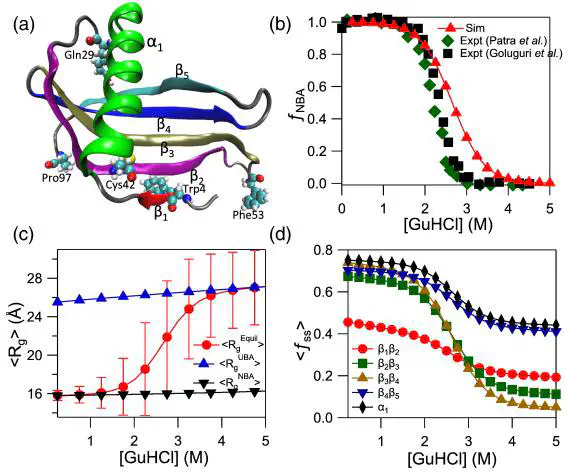 Image credit: G. Reddy
Image credit: G. Reddy
Abstract
Proteins, which behave as random coils in high denaturant concentrations undergo collapse transition similar to polymers on denaturant dilution. We study collapse in the denatured ensemble of single-chain monellin (MNEI) using a coarse-grained protein model and molecular dynamics simulations. The model is validated by quantitatively comparing the computed guanidinium chloride and pH-dependent thermodynamic properties of MNEI folding with the experiments. The computed properties such as the fraction of the protein in the folded state and radius of gyration (Rg) as function of [GuHCl] are in good agreement with the experiments. The folded state of MNEI is destabilized with an increase in pH due to the deprotonation of the residues Glu24 and Cys42. On decreasing [GuHCl], the protein in the unfolded ensemble showed specific compaction. The Rg of the protein decreased steadily with [GuHCl] dilution due to increase in the number of native contacts in all the secondary structural elements present in the protein. MNEI folding kinetics is complex with multiple folding pathways and transiently stable intermediates are populated in these pathways. In strong stabilizing conditions, the protein in the unfolded ensemble showed transition to a more compact unfolded state where Rg decreased by ≈ 17% due to the formation of specific native contacts in the protein. The intermediate populated in the dominant MNEI folding pathway satisfies the structural features of the dry molten globule inferred from experiments