 Image credit: G. Reddy
Image credit: G. Reddy
Abstract
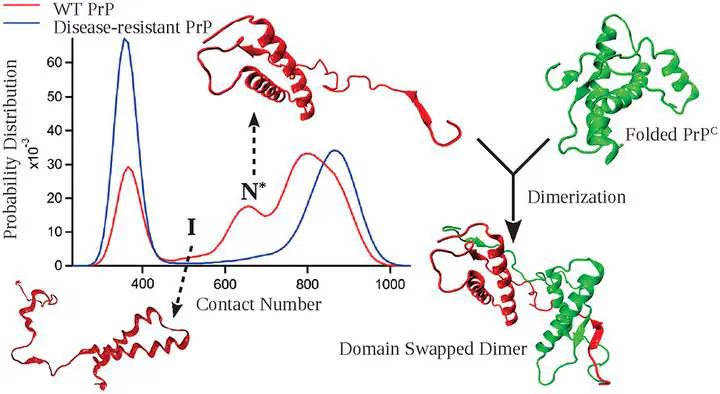
Aggregation of misfolded prion proteins causes fatal neurodegenerative disorders in both humans and animals. There is an extensive effort to identify the elusive aggregation-prone conformations (N*) of prions, which are early stage precursors to aggregation. We studied temperature- and force-induced unfolding of the structured C-terminal domain of mouse (moPrP) and human prion proteins (hPrP) using molecular dynamics simulations and coarse-grained protein models. We find that these proteins sparsely populate intermediate states bearing the features of N* and readily undergo domain-swapped dimerization by swapping the short β-strands present at the beginning of the C-terminal domain. The structure of the N* state is similar for both moPrP and hPrP, indicating a common pathogenic precursor across different species. Interestingly, disease-resistant hPrP (G127V) showed a drastic reduction in the population of the N* state further hinting a pathogenic connection to these partially denatured conformations. This study proposes a plausible runaway domain-swapping mechanism to describe the onset of prion aggregation.