 Image credit: G. Reddy
Image credit: G. Reddy
Abstract
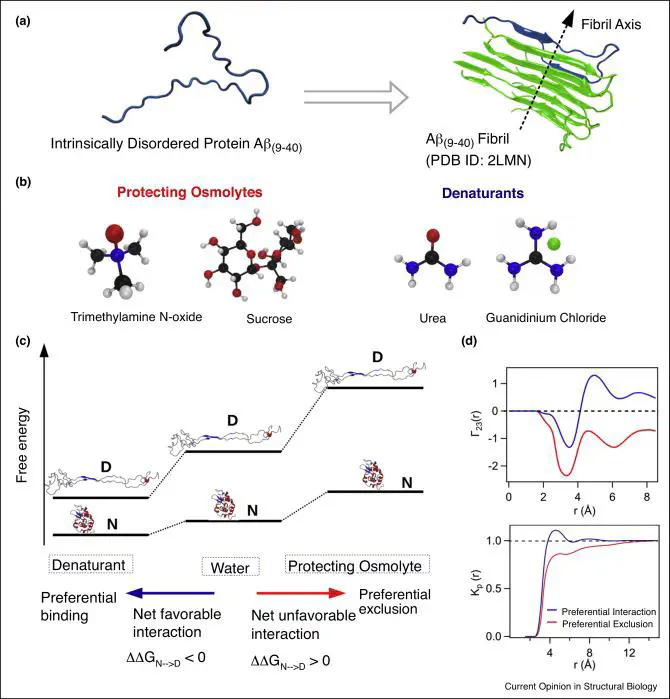
Cells are equipped with cosolvents that modulate proteinfolding and aggregation to withstand water stress. The effect ofcosolvents on the aggregation rates depends on whether thepolypeptide sequence is an intrinsically disordered protein(IDP) or can fold into a specific native structure. Cosolvents,which act as denaturants generally slow down aggregation inIDPs, while expediting it in globular proteins. In contrast,protecting osmolytes facilitate aggregation in IDPs, whileslowing it down in globular proteins. In this review we highlightthe recent computational approaches to gain insight into therole of cosolvents on the aggregation mechanism of IDPs andglobular proteins. Computer simulations using the moleculartransfer model, which implements the cosolvent effects incoarse-grained protein models in conjunction with enhancedsampling techniques played an important role in elucidating theeffect of cosolvents on the growth of amyloid fibrils.